A powerful Software for Molecular Visualization.
From open source to pro services, Furious Atoms helps you to visualize, build, simulates molecular systems.
From open source to pro services, Furious Atoms helps you to visualize, build, simulates molecular systems.

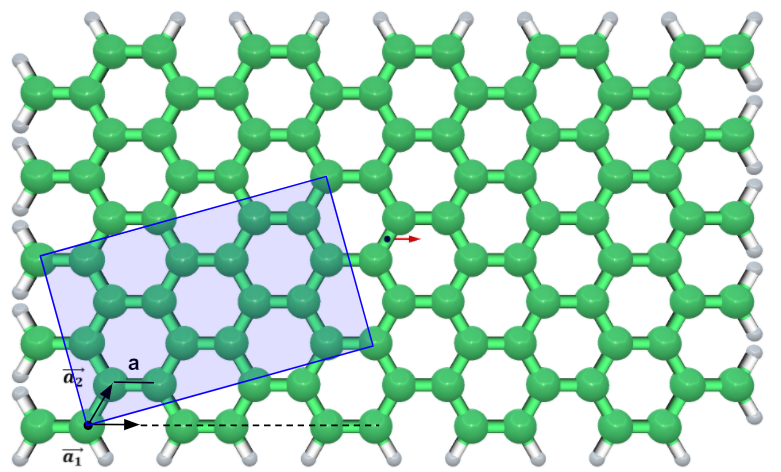
Graphene Sheet feature allows construction of graphene and graphene-like sheets such as hexagonal boron nitride (h-BN), boron phosphide (h-BP), hexagonal aluminum phosphonate (h-AlP) and hexagonal aluminum nitride (h-AlN). The Graphene Sheet is located in the Build menu under Build Nanostructures. Graphene is made of a single atomic layer. On the graphene net, the chiral indices (n, m) specify the lattice structure. n and m are numerically positive integers and define the chiral vector ($\overrightarrow{C_{h}}$) as: \begin{equation} \overrightarrow{C_{h}}=(n, m)=n \overrightarrow{a_{1}}+m \overrightarrow{a_{2}} \end{equation} Where $\overrightarrow{a_{1}}$ and $\overrightarrow{a_{2}}$ are primitive lattice vectors in the hexagonal lattice of graphene separated with an inner angle of 60. a is the bond length of c-c (the distance between nearest neighbors), the primitive lattice vectors can be chosen to be: \begin{equation} \vec{a}_{1}=\frac{a}{2}(3, \sqrt{3}), \vec{a}_{2}=\frac{a}{2}(3,-\sqrt{3}) \end{equation} Based on chiral indices (n and m), graphene can be further categorized into “armchair” when n=m, “zig-zag” when n > 0, m = 0, and all other integer combinations are called “chiral”. The default type of sheet structure is carbon-carbon (C-C) with bond length of 1.421 $\text { Å }$. Figure 1. The graphene lattice geometry. a1 and a2 are the lattice unit vectors. a is the bond length of carbon carbon (c-c). The user can change the bond length of graphene and graphene-like sheets. In the “Hydrogen termination” option, the edges of the structure are terminated by hydrogen, if "All" is selected.
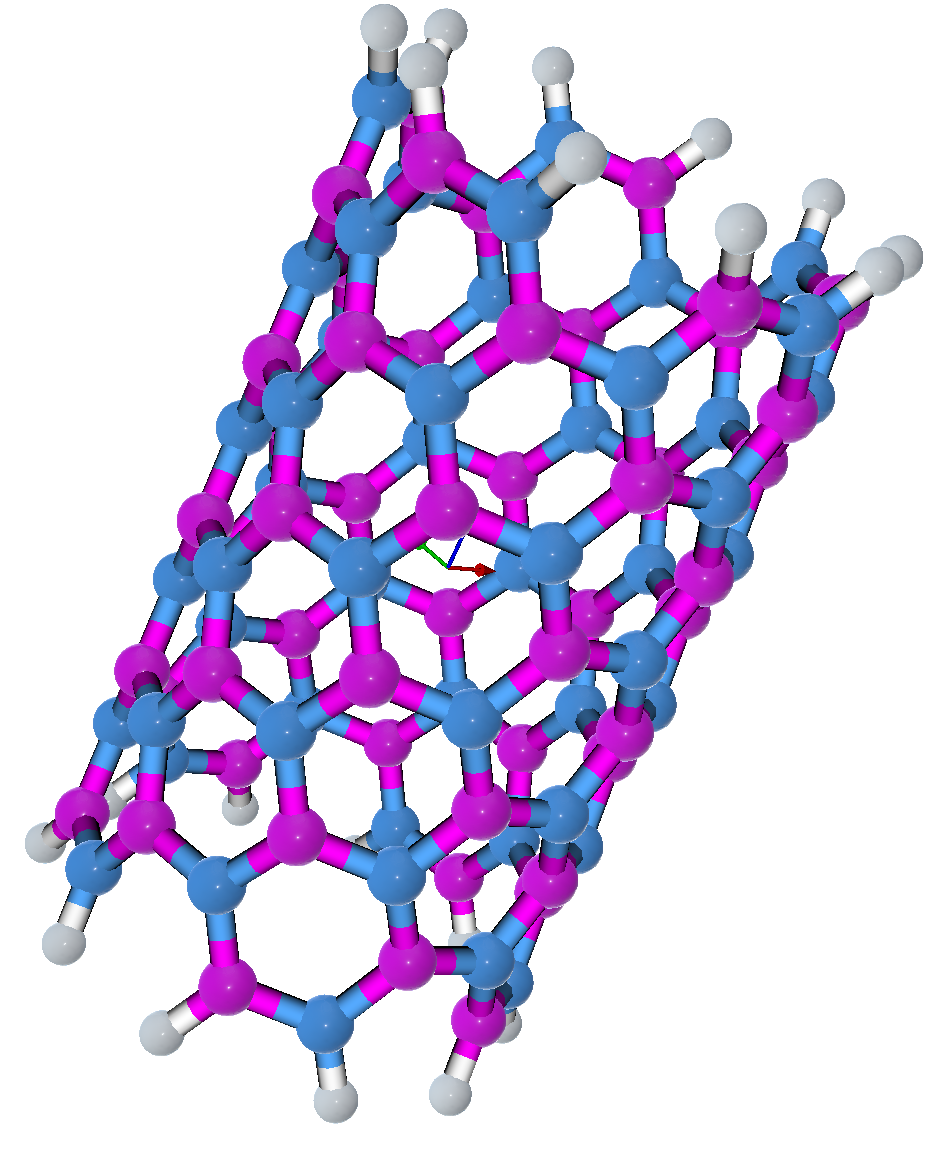
The Single-Walled Nanotube (SWNT) builder is located under the Build menu. It generates SWNT by first creating a flat graphene or graphene-like lattice with chiral indices n and m provided by the user. This hexagonal lattice is then rolled onto a cylinder to create the SWNT unit cell aligned in the z-direction. The diameter (d) of SWNT is defined as: \begin{equation} \begin{aligned} &d=\frac{A}{\pi}=\frac{a_{0}}{\pi} \sqrt{n^{2}+m^{2}+n m} \\ &d=|\mathbf{C}| / p i=\operatorname{asqrt}\left(\mathrm{n}^{2}+\mathrm{n} \mathrm{m}+\mathrm{m}^{2}\right) \end{aligned} \end{equation} Where a is the lattice constant. For graphene, a = 2.49 A. The length of the SWCN unit cell can be written as: \begin{equation} |\mathrm{T}|=\operatorname{sqrt}(3) d / \operatorname{gcd}(2 m+n, m+2 n) \end{equation} The user can build hexagonal boron nitride nanotube (BNNT), hexagonal boron phosphide nanotube (BPNT), hexagonal aluminum phosphonate nanotube (AlPNT) and hexagonal aluminum nitride nanotube (AlNNT) in form of single wall structures. You can set the n, m indexes, atom types, bond length, repeat units to determine the type of nanotube. All unit distances are in angstrom. In the “Hydrogen termination” option, you can select “One end” or “Both ends” to terminate one edge or both edges of the nanotube with hydrogen respectively. The information about the diameter and the length of the nanotube is updated in the widget while the user selects the n, m and repeat unit parameters. You can save the generated file into XYZ ( for example: SWNT.xyz), PDB ( for example: SWNT.pdb), LAMMPS, (for example: SWNT.data) and GROMACS format (for example: SWNT.gro).
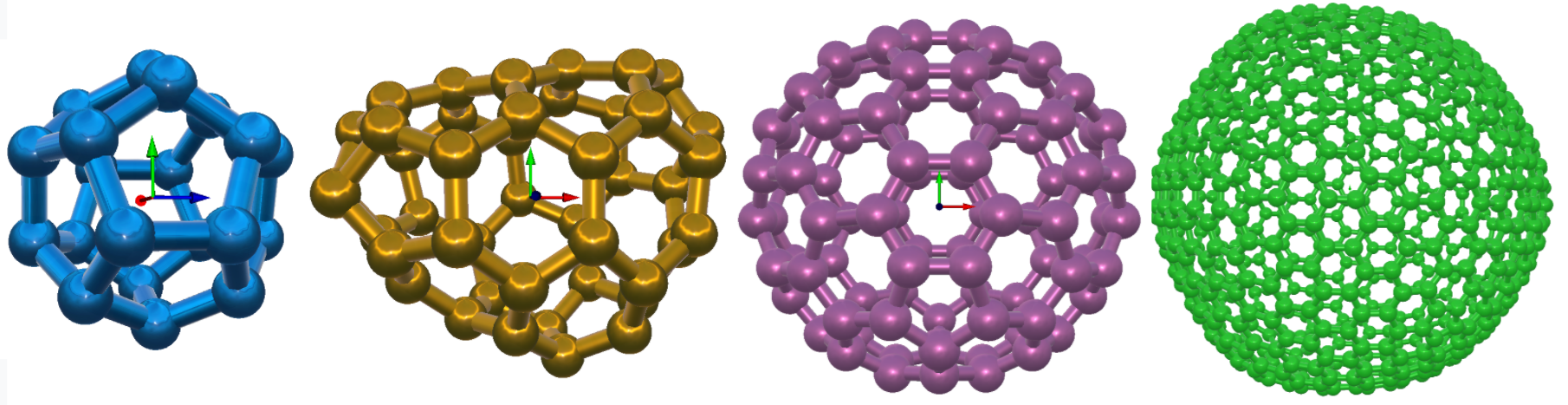
The Multi-Walled Nanotube (MWNT) builder is located under the Build menu. It generates multiple nanotubes with wall separation of 3.43 A aligned along the z-axis. You can choose the indices n and m (to define the type of chirality), type of the atoms, bond length, repeat units (to define the length of MWNT) and the number of walls in MWNT. All unit distances are in angstrom. The user can see the updated values of diameter inner nanotube and the length of MWNT as the n, m and repeat unit parameters are changing in the widget. Like SWNT, in MWNT the user can build hexagonal boron nitride nanotube (BNNT), hexagonal boron phosphide nanotube (BPNT), hexagonal aluminum phosphonate nanotube (AlPNT) and hexagonal aluminum nitride nanotube (AlNNT) in the form of multiple walls structures. You can save the generated file into XYZ ( for example: MWNT.xyz), PDB ( for example: MWNT.pdb), LAMMPS, (for example: MWNT.data) and GROMACS format (for example: MWNT.gro).

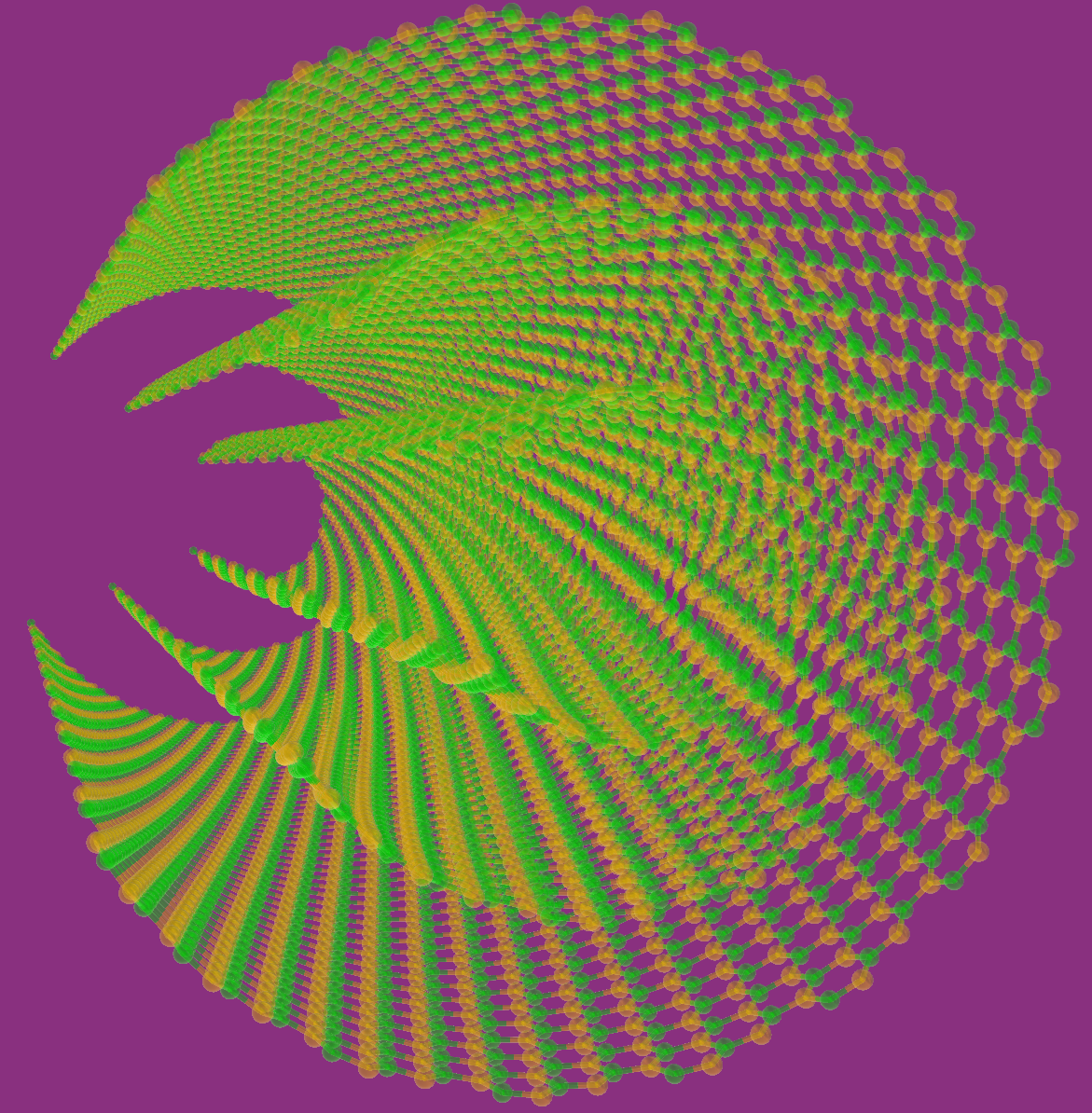
The Fullerene builder is located under the Build menu. With Fullerene Builder, the user can create more than 2450 derivatives of fullerene. The user can save the generated file into XYZ ( for example: fullerene.xyz), PDB ( for example: graphene.pdb), LAMMPS, (for example: fullerene.data) and GROMACS format (for example: fullerene.gro).
The Electrolyte builder is located under the Build menu. This feature enables the users to create different types of electrolytes for All-Atom (with water molecules) and Coarse-grained (without water molecules) Molecular Dynamics Simulations. These models can be confined inside the nanoscale channels by enabling the “create dummy wall in z direction” icon. There are three steps to build the electrolyte.

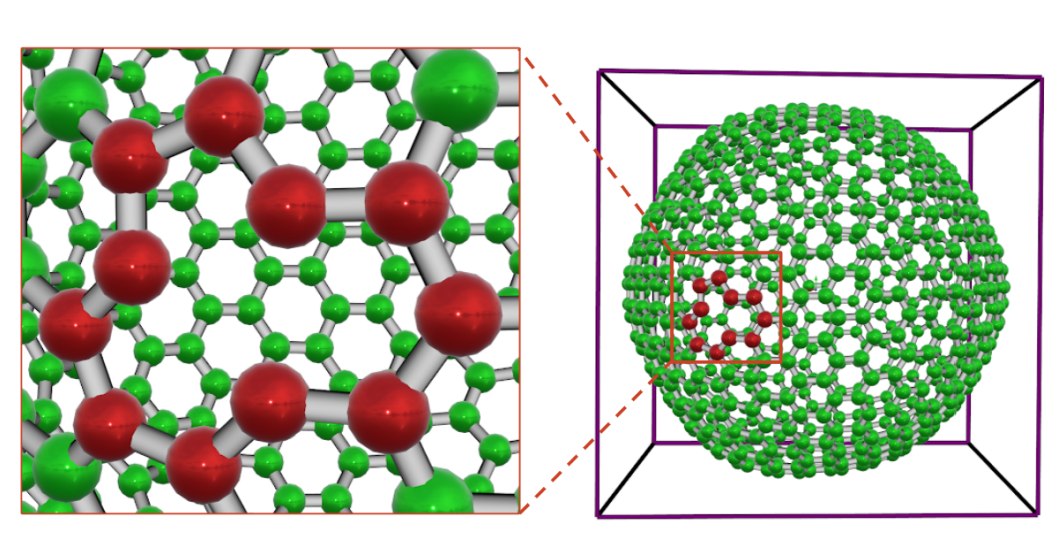
FA allows the users to modify the chemical structures by selecting the atoms or bonds and deleting them. The user can also add a simulation box to the model or change the size of the existing simulation box. As an example, we presented a fullerene C720 that is built with FA. The user can easily edit C720 and create a pristine single vacancy (SV) on the cage by removing one carbon atom and simultaneously breaking three C-C bonds in the network. The carbons that are involved with the defected region are shown in red. Moreover, the users can convert the Coarse-Grained model to an All-Atom model by adding the water model into the selected system. After modification, the structure can be saved and exported to LAMMPS and PDB file formats.
FURIOUS ATOMS uses the FURY visualization library to design and modify the size, shape, color and positions of structures while observing the changes in real time.
Please Cite:
All the basics steps to start using Furious Atoms
All the basics steps for contributing in Furious Atoms
A. FURIOUS ATOMS uses the FURY visualization library to design and modify the size, shape, color and positions of structures while observing the changes in real time. Please Cite:
1. Eleftherios Garyfallidis, Serge Koudoro, Javier Guaje, Marc-Alex Côté, Soham Biswas, David Reagan, Nasim Anousheh, Filipi Silva, Geoffrey Fox, and Fury Contributors. "FURY: advanced scientific visualization." Journal of Open Source Software 6, no. 64 (2021): 3384.A. Furious Atoms allows researchers to build chemical structures of interest in 3D interactively. The users can generate the nanostructures such as graphene, Single-Walled Nanotubes, Multi-walled Nanotubes, Fullerenes (more than 2450 derivatives) and electrolyte models in both All-Atom and Coarse-Grained models. The software can generate Boron-Nitride (BN), Aluminum-Nitride (AlN) and Gallium-Nitride (GaN) nanotubes and graphene-like structures.
A. Furious Atoms enables chemists to build chemical structures from the ground up or modify existing structures (by deleting atoms and bonds) which can then be saved into common file formats such as LAMMPS, Protein Data Bank (PDB), GROMACS and xyz format.